library(tidyverse)
library(cowplot)
library(broom)
library(modelr)
library(viridis)
library(lubridate)
library(hms)
library(knitr)
library(kableExtra)
knitr::opts_chunk$set(tidy.opts=list(width.cutoff=60),tidy=TRUE, echo = TRUE, message=FALSE, warning=FALSE, fig.align="center")
source("../../tools/echem_processing_tools.R")
source("../../tools/plotting_tools.R")
theme_set(theme_1())
soak_1_path = "../data/soak_1/"
tran_1_path = "../data/transfer_1/"
data_cols <- c("E", "i1", "i2")
swv_skip_rows = 18
gc_skip_rows = 21
All SWVs
# Add 'reactor' to file name so it is parsed into column
filename_cols = c("reactor", "echem", "rep")
swv_tran_1_names <- dir(path = tran_1_path, pattern = "[swv]+.+[txt]$") %>%
paste("tran1", ., sep = "_")
swv_soak_1_names <- dir(path = soak_1_path, pattern = "[swv]+.+[txt]$") %>%
paste("soak1", ., sep = "_")
# Add correct paths separate from filenames
swv_tran_1_paths <- dir(path = tran_1_path, pattern = "[swv]+.+[txt]$") %>%
paste(tran_1_path, ., sep = "")
swv_soak_1_paths <- dir(path = soak_1_path, pattern = "[swv]+.+[txt]$") %>%
paste(soak_1_path, ., sep = "")
# Combine all SWVs into single vector
swv_names <- c(swv_tran_1_names, swv_soak_1_names)
swv_paths <- c(swv_tran_1_paths, swv_soak_1_paths)
# Read in all SWVs with one function call
swv_data <- echem_import_to_df(filenames = swv_names, file_paths = swv_paths,
data_cols = data_cols, skip_rows = swv_skip_rows, filename_cols = filename_cols,
rep = T, PHZadded = F)
swv_data %>% head() %>% kable() %>% kable_styling()
|
reactor
|
echem
|
rep
|
minutes
|
E
|
electrode
|
current
|
|
tran1
|
swv
|
1
|
1334.583
|
0.099
|
i1
|
2.1e-06
|
|
tran1
|
swv
|
1
|
1334.583
|
0.098
|
i1
|
3.1e-06
|
|
tran1
|
swv
|
1
|
1334.583
|
0.097
|
i1
|
3.1e-06
|
|
tran1
|
swv
|
1
|
1334.583
|
0.096
|
i1
|
3.1e-06
|
|
tran1
|
swv
|
1
|
1334.583
|
0.095
|
i1
|
3.1e-06
|
|
tran1
|
swv
|
1
|
1334.583
|
0.094
|
i1
|
3.1e-06
|
swv_data %>% filter(reactor == "soak1") %>% ggplot(., aes(x = E,
y = current, color = rep, group = rep)) + geom_path() + facet_wrap(~electrode,
scales = "free") + scale_color_viridis() + scale_x_reverse()
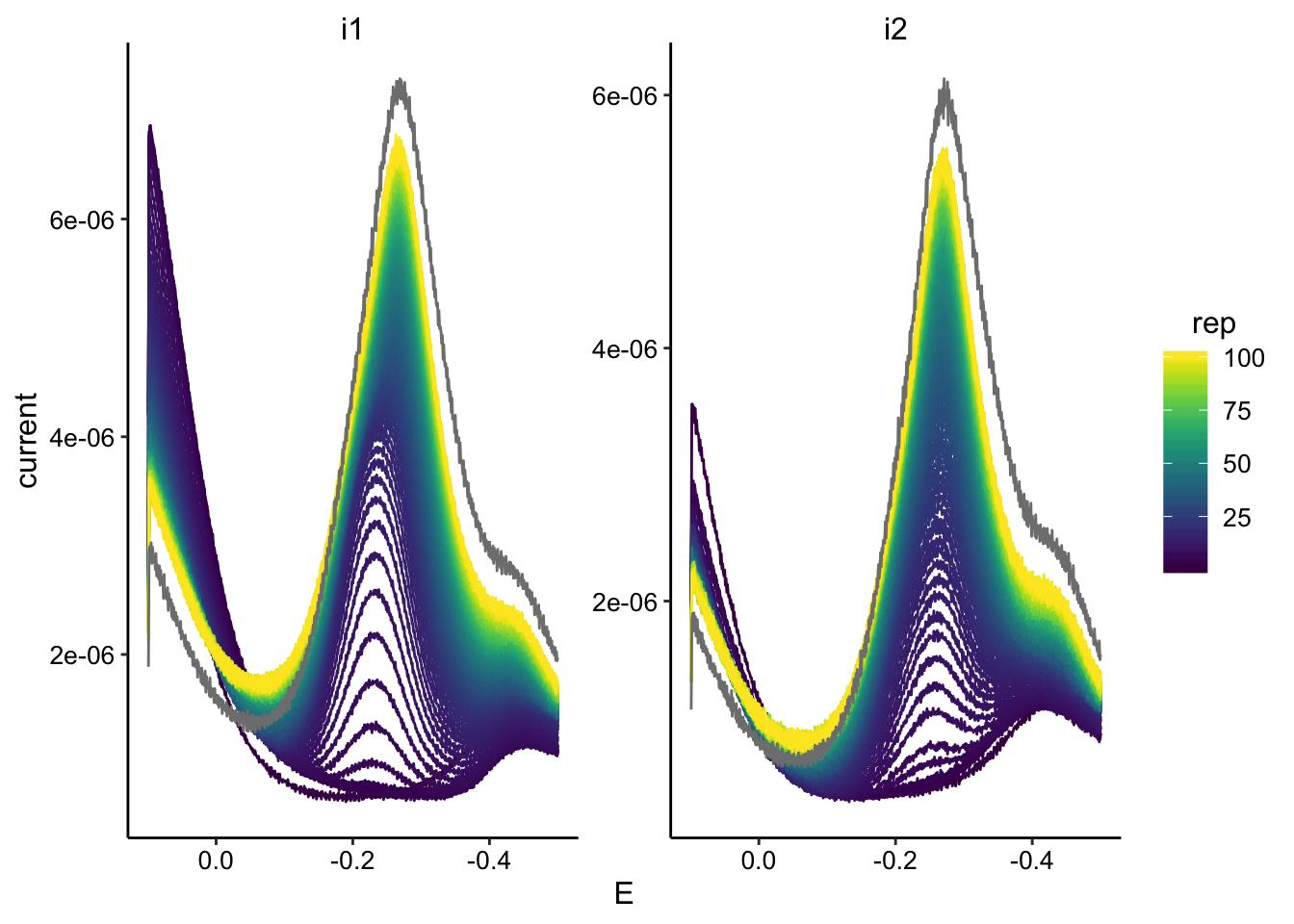
swv_data %>% filter(reactor == "tran1") %>% ggplot(., aes(x = E,
y = current, color = rep, group = rep)) + geom_path() + facet_wrap(~electrode,
scales = "free") + scale_color_viridis() + scale_x_reverse()
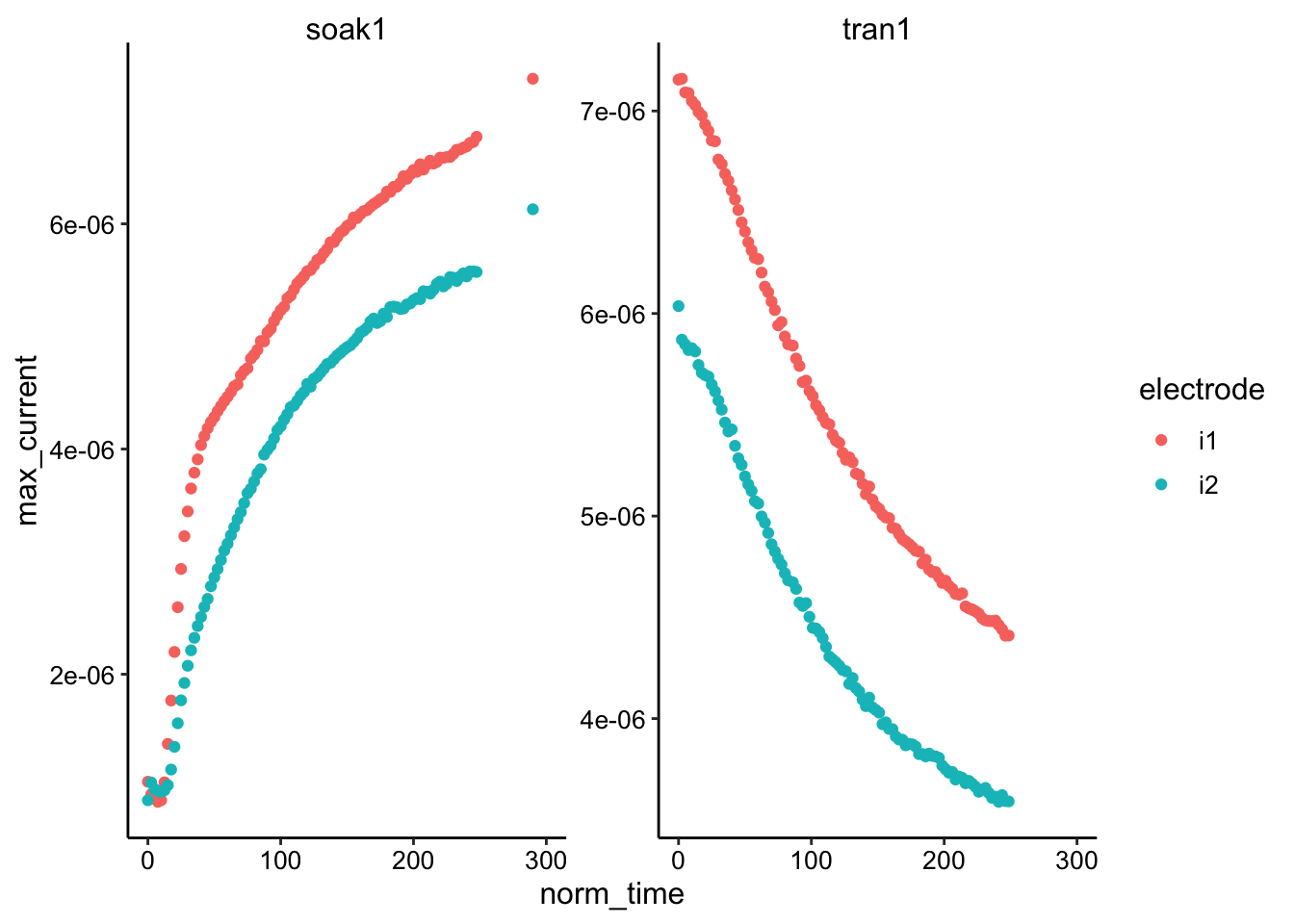
swv_max <- swv_data %>% mutate(minutes = ifelse(minutes < 1000,
minutes + 1440, minutes)) %>%
group_by(reactor) %>% mutate(min_time = min(minutes)) %>% mutate(norm_time = minutes -
min_time) %>%
group_by(reactor, rep, electrode) %>% filter(E < -0.15 & E >
-0.35) %>% mutate(max_current = max(abs(current))) %>% filter(abs(current) ==
max_current)
ggplot(swv_max, aes(x = norm_time, y = max_current, color = electrode)) +
geom_point() + facet_wrap(~reactor, scales = "free") + xlim(0,
300)
All GCs
# Add 'reactor' to file name so it is parsed into column
filename_cols = c("reactor", "echem", "rep")
gc_tran_1_names <- dir(path = tran_1_path, pattern = "[gc]+.+[txt]$") %>%
paste("tran1", ., sep = "_")
gc_soak_1_names <- dir(path = soak_1_path, pattern = "[gc]+.+[txt]$") %>%
paste("soak1", ., sep = "_")
# Add correct paths separate from filenames
gc_tran_1_paths <- dir(path = tran_1_path, pattern = "[gc]+.+[txt]$") %>%
paste(tran_1_path, ., sep = "")
gc_soak_1_paths <- dir(path = soak_1_path, pattern = "[gc]+.+[txt]$") %>%
paste(soak_1_path, ., sep = "")
# Combine all SWVs into single vector
gc_names <- c(gc_tran_1_names, gc_soak_1_names)
gc_paths <- c(gc_tran_1_paths, gc_soak_1_paths)
# Read in all SWVs with one function call
gc_data <- echem_import_to_df(filenames = gc_names, file_paths = gc_paths,
data_cols = data_cols, skip_rows = gc_skip_rows, filename_cols = filename_cols,
rep = T, PHZadded = F)
gc_data %>% head() %>% kable(digits = 9) %>% kable_styling()
|
reactor
|
echem
|
rep
|
minutes
|
E
|
electrode
|
current
|
|
tran1
|
gc
|
1
|
1336.9
|
0.000
|
i1
|
-4.1e-08
|
|
tran1
|
gc
|
1
|
1336.9
|
-0.001
|
i1
|
-3.5e-08
|
|
tran1
|
gc
|
1
|
1336.9
|
-0.002
|
i1
|
-3.3e-08
|
|
tran1
|
gc
|
1
|
1336.9
|
-0.003
|
i1
|
-2.8e-08
|
|
tran1
|
gc
|
1
|
1336.9
|
-0.004
|
i1
|
-2.7e-08
|
|
tran1
|
gc
|
1
|
1336.9
|
-0.005
|
i1
|
-2.3e-08
|
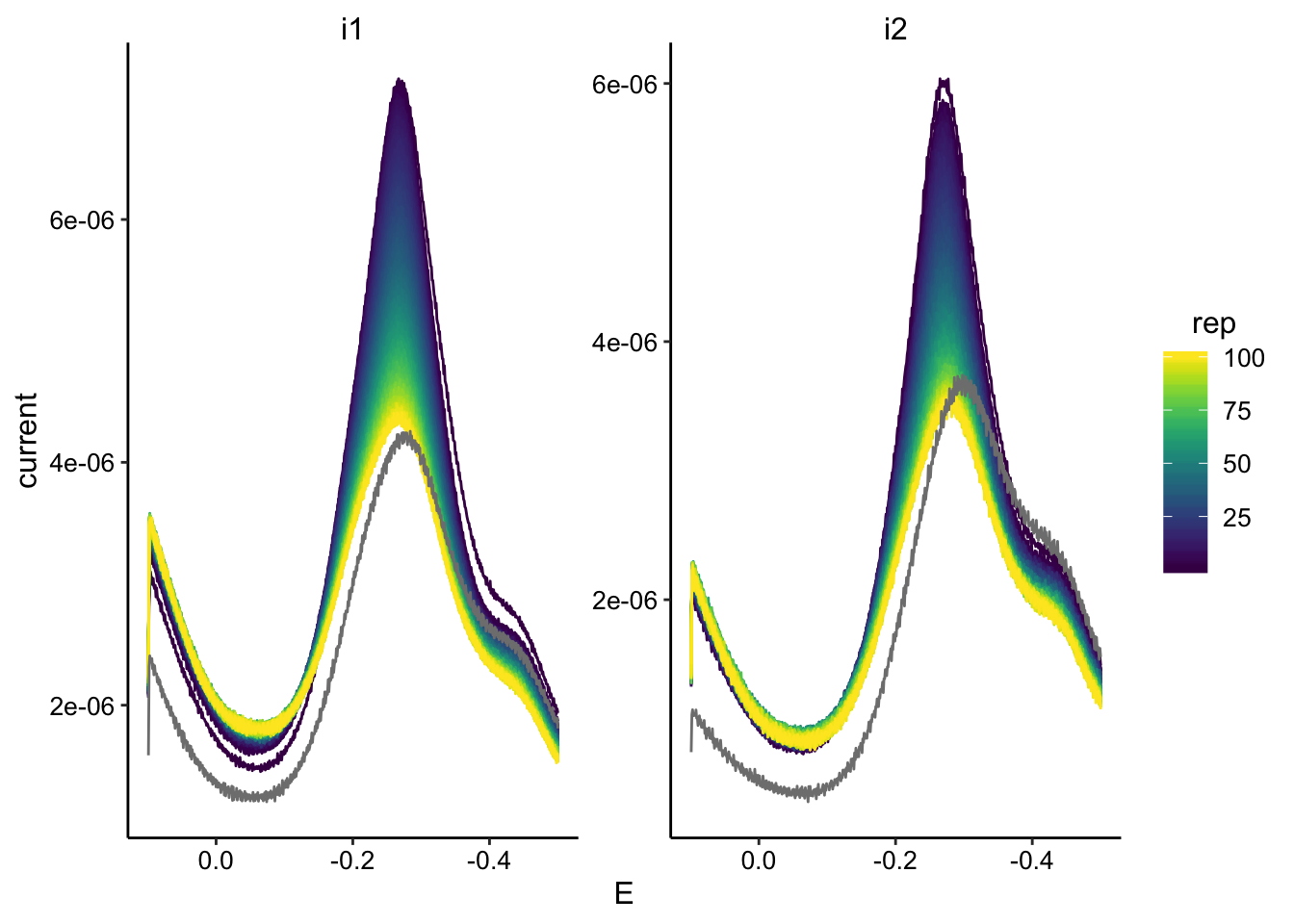
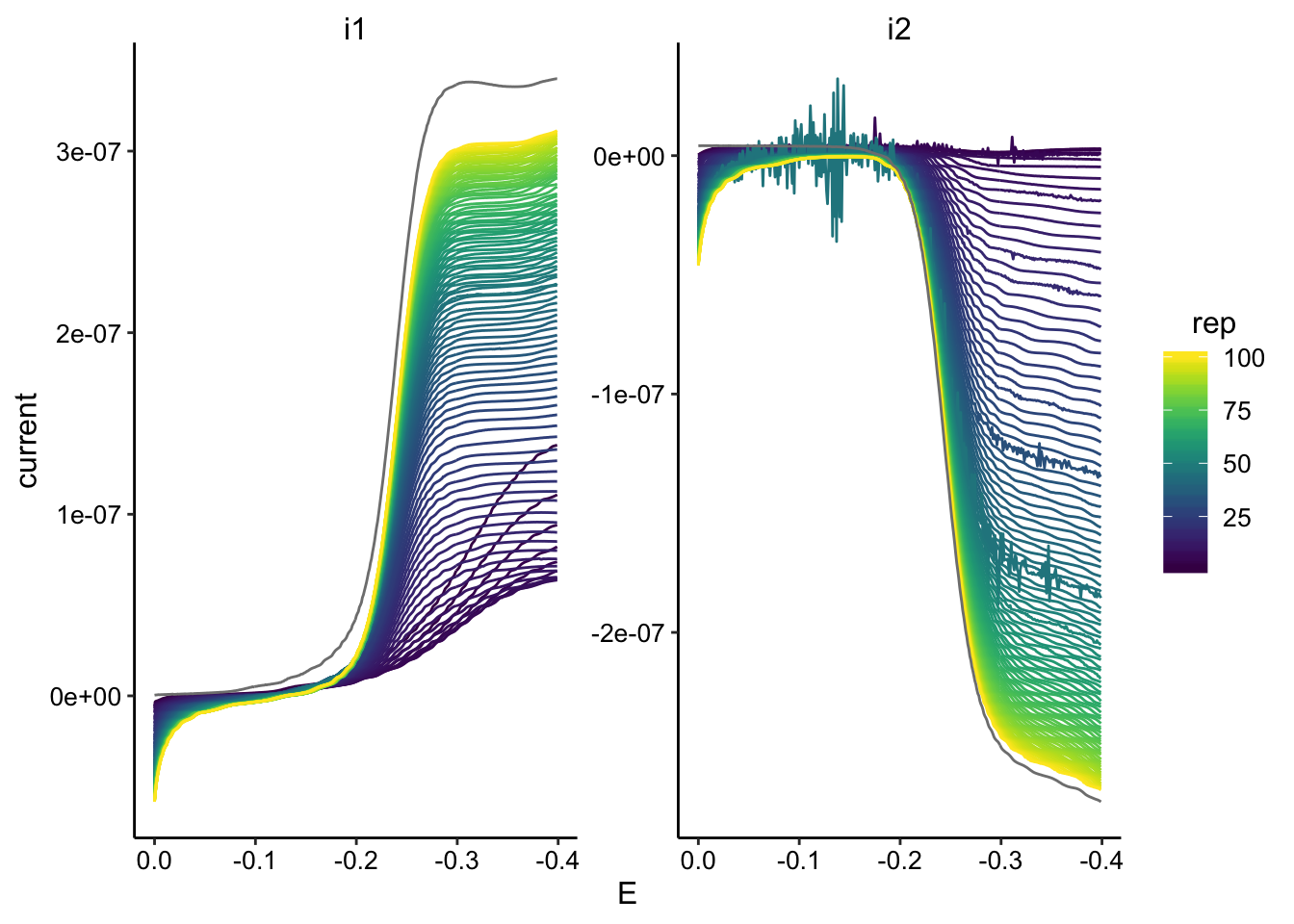
gc_data %>% filter(reactor == "soak1") %>% ggplot(., aes(x = E,
y = current, color = rep, group = rep)) + geom_path() + facet_wrap(~electrode,
scales = "free") + scale_color_viridis() + scale_x_reverse()
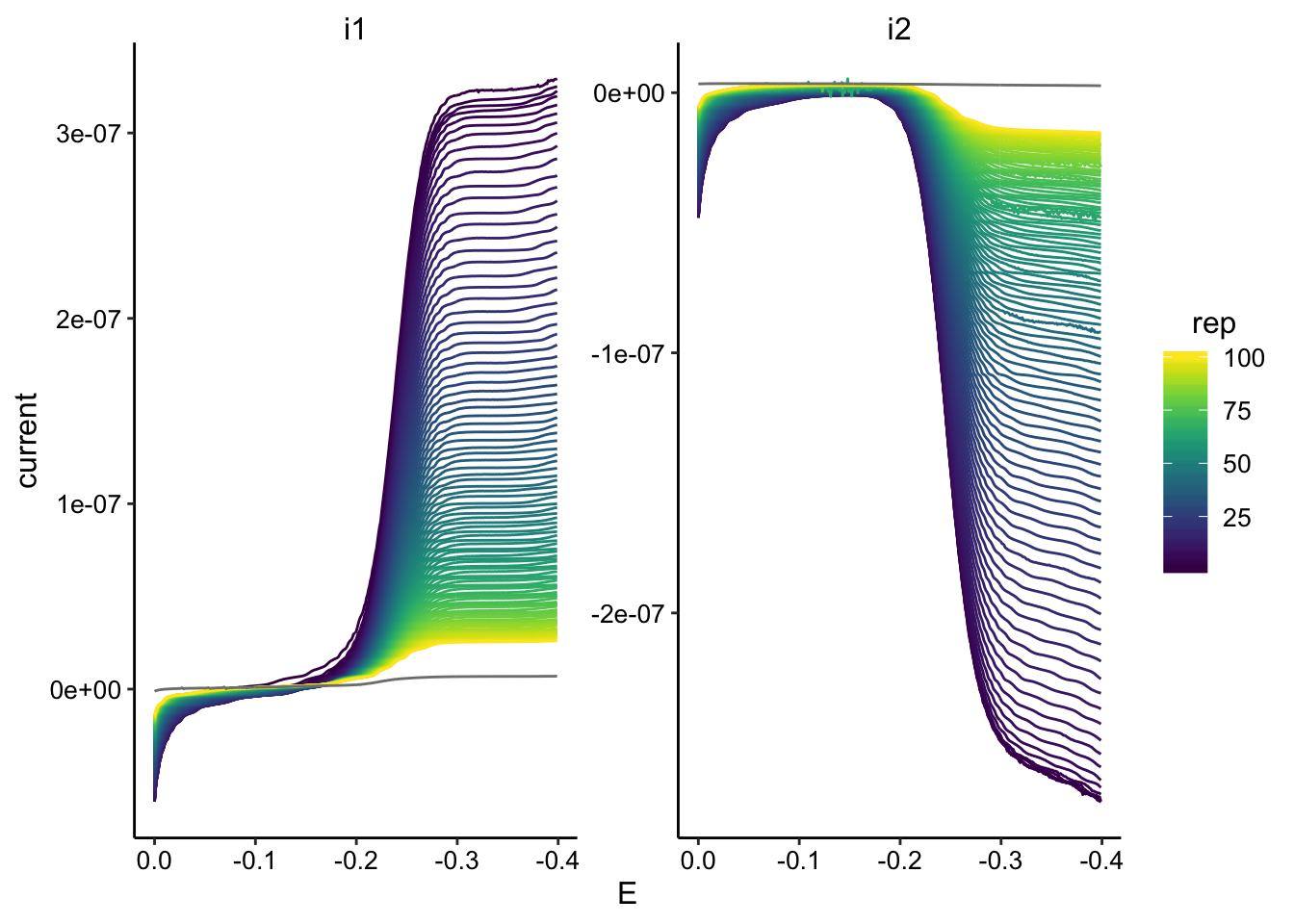
gc_data %>% filter(reactor == "tran1") %>% ggplot(., aes(x = E,
y = current, color = rep, group = rep)) + geom_path() + facet_wrap(~electrode,
scales = "free") + scale_color_viridis() + scale_x_reverse()
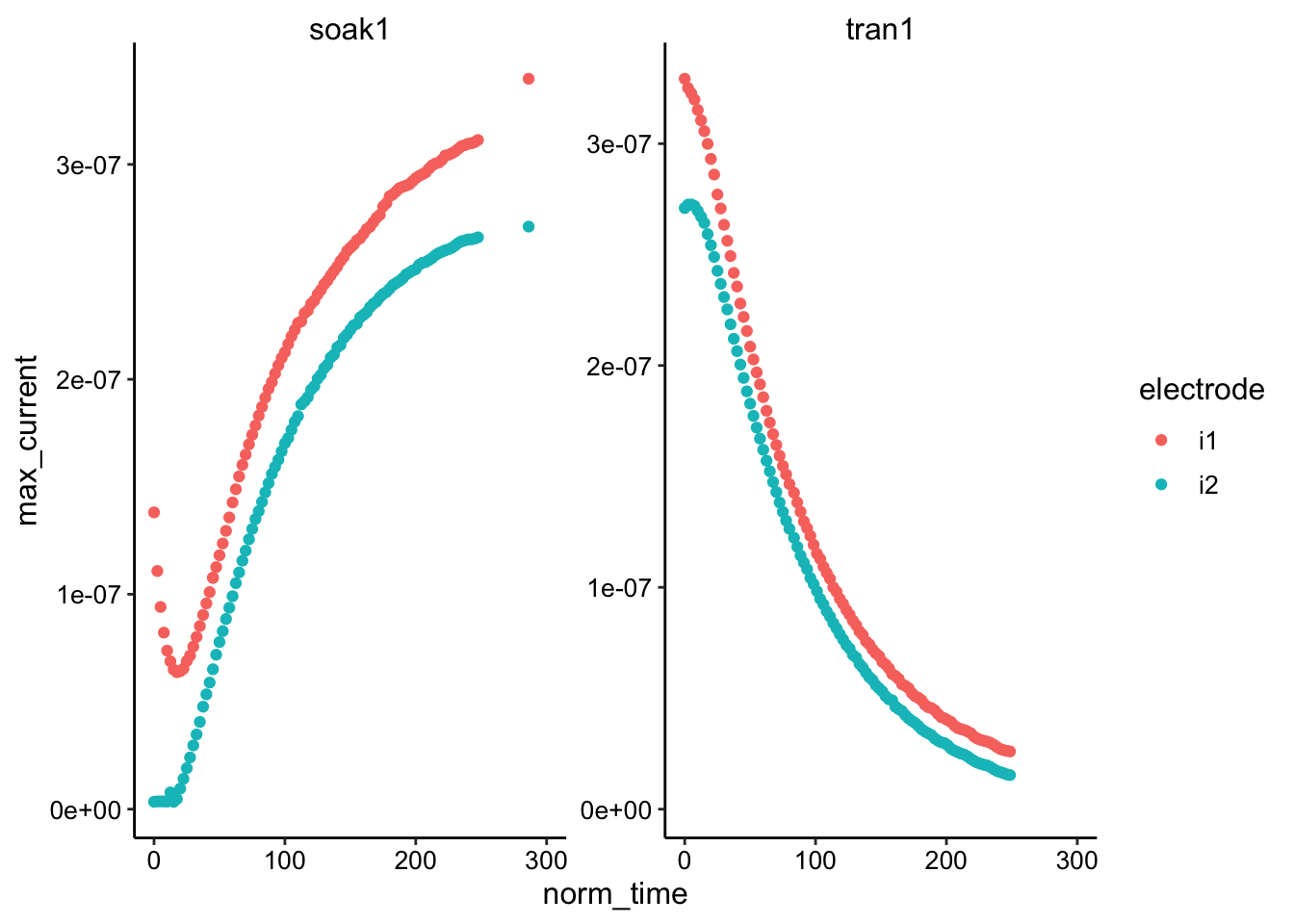
gc_max <- gc_data %>% mutate(minutes = ifelse(minutes < 1000,
minutes + 1440, minutes)) %>%
group_by(reactor) %>% mutate(min_time = min(minutes)) %>% mutate(norm_time = minutes -
min_time) %>%
group_by(reactor, rep, electrode) %>% filter(E < -0.2) %>% mutate(max_current = max(abs(current))) %>%
filter(abs(current) == max_current)
ggplot(gc_max, aes(x = norm_time, y = max_current, color = electrode)) +
geom_point() + facet_wrap(~reactor, scales = "free") + xlim(0,
300)
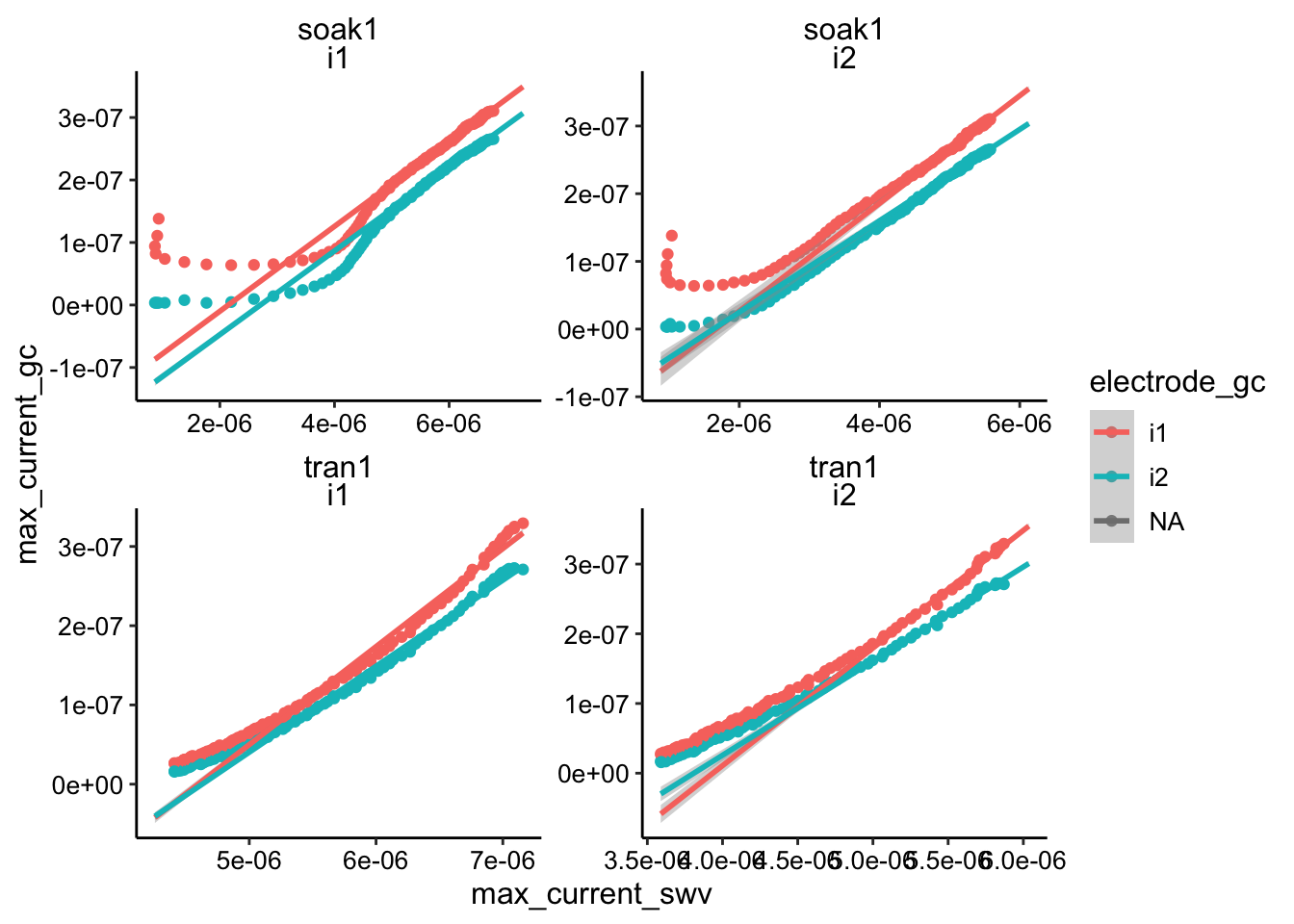
SWV vs. GC
swv_gc_max <- left_join(swv_max %>% ungroup() %>% mutate(rep = rep -
1), gc_max, by = c("reactor", "rep"), suffix = c("_swv",
"_gc"))
ggplot(swv_gc_max, aes(x = max_current_swv, y = max_current_gc,
color = electrode_gc)) + geom_point() + geom_smooth(data = swv_gc_max %>%
filter(max_current_swv > 5e-06), method = "lm", fullrange = T) +
facet_wrap(c("reactor", "electrode_swv"), scales = "free")
ggplot(swv_gc_max %>% filter(electrode_gc == "i2"), aes(x = max_current_swv,
y = max_current_gc, color = reactor, shape = electrode_swv)) +
geom_point()
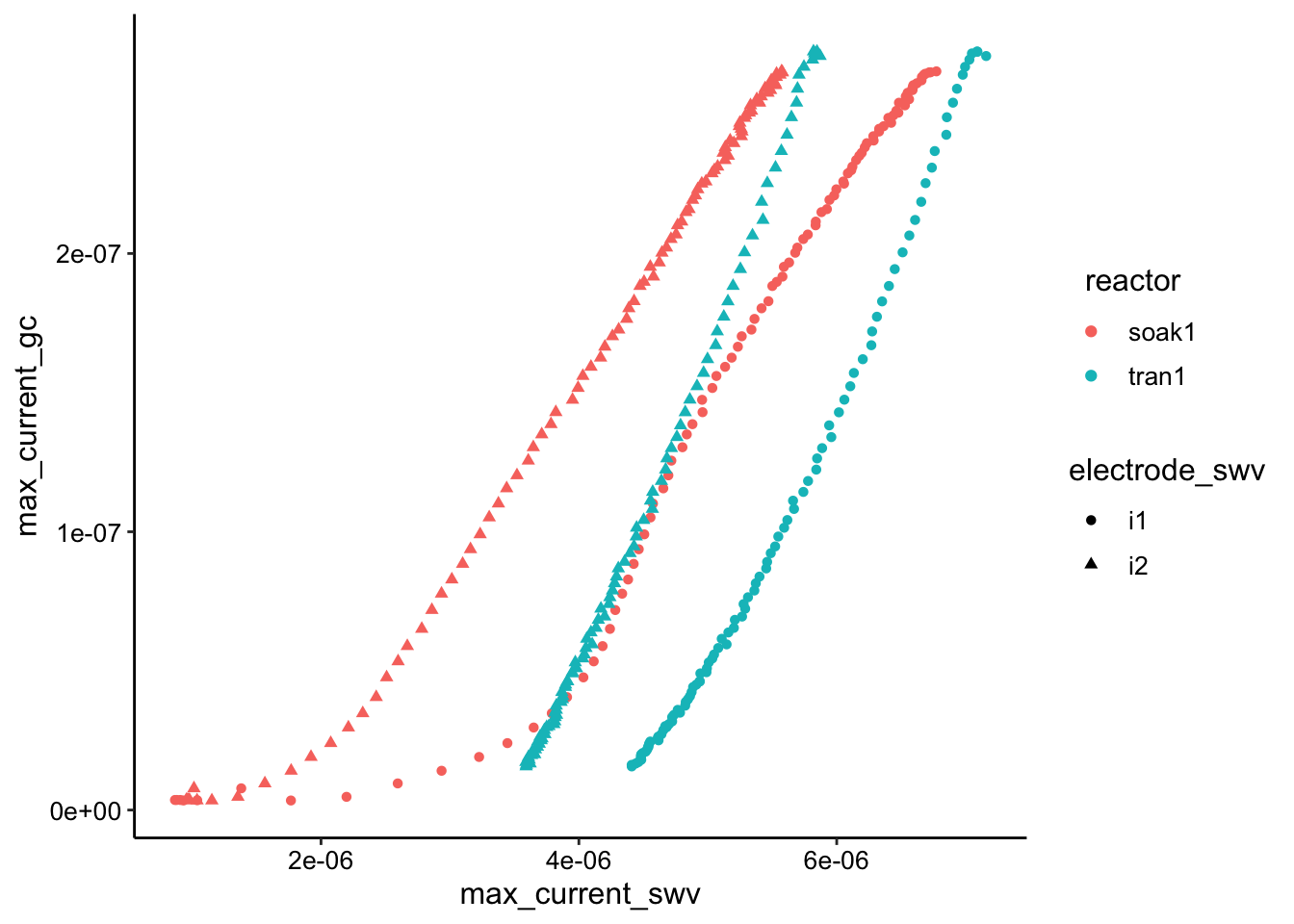
ggplot(swv_gc_max %>% filter(electrode_gc == "i2" & reactor ==
"soak1"), aes(x = max_current_swv, y = max_current_gc, color = reactor,
shape = electrode_swv)) + geom_point() + geom_smooth(data = swv_gc_max %>%
filter(electrode_gc == "i2" & reactor == "soak1" & rep >
30), method = "lm", fullrange = T)
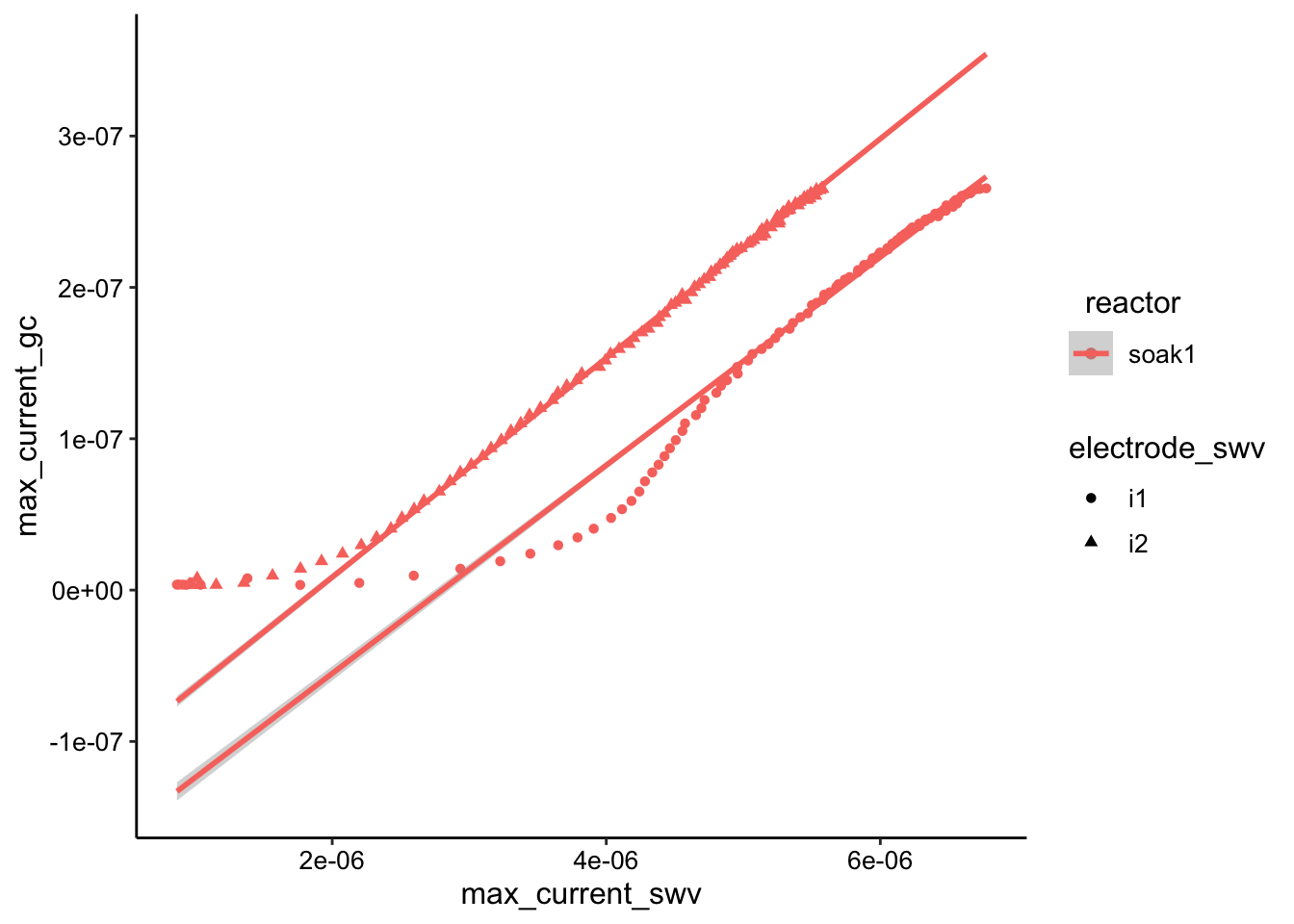
ggplot(swv_gc_max %>% filter(electrode_gc == "i2" & reactor ==
"tran1"), aes(x = max_current_swv, y = max_current_gc, color = reactor,
shape = electrode_swv)) + geom_point() + geom_smooth(data = swv_gc_max %>%
filter(electrode_gc == "i2" & reactor == "tran1" & rep <
50), method = "lm", fullrange = T)
dap_from_swvGC <- function(m, t_p = 1/(2 * 300)) {
psi <- 0.7
# psi <- 0.75 A <- 0.013 #cm^2
A <- 0.025 #cm^2
S <- 18.4 #cm
d_ap <- (m * A * psi)^2/(S^2 * pi * t_p)
d_ap
}
lms <- swv_gc_max %>% filter(rep > 0 & max_current_swv > 5e-06) %>%
group_by(reactor, electrode_swv, electrode_gc) %>% do(tidy(lm(max_current_gc ~
max_current_swv, data = .), conf.int = T)) %>% filter(term ==
"max_current_swv") %>% mutate(dap = dap_from_swvGC(m = estimate)) %>%
mutate(dap_high = dap_from_swvGC(m = conf.high)) %>% mutate(dap_low = dap_from_swvGC(m = conf.low)) %>%
mutate(dataset = "SWVvsGC")
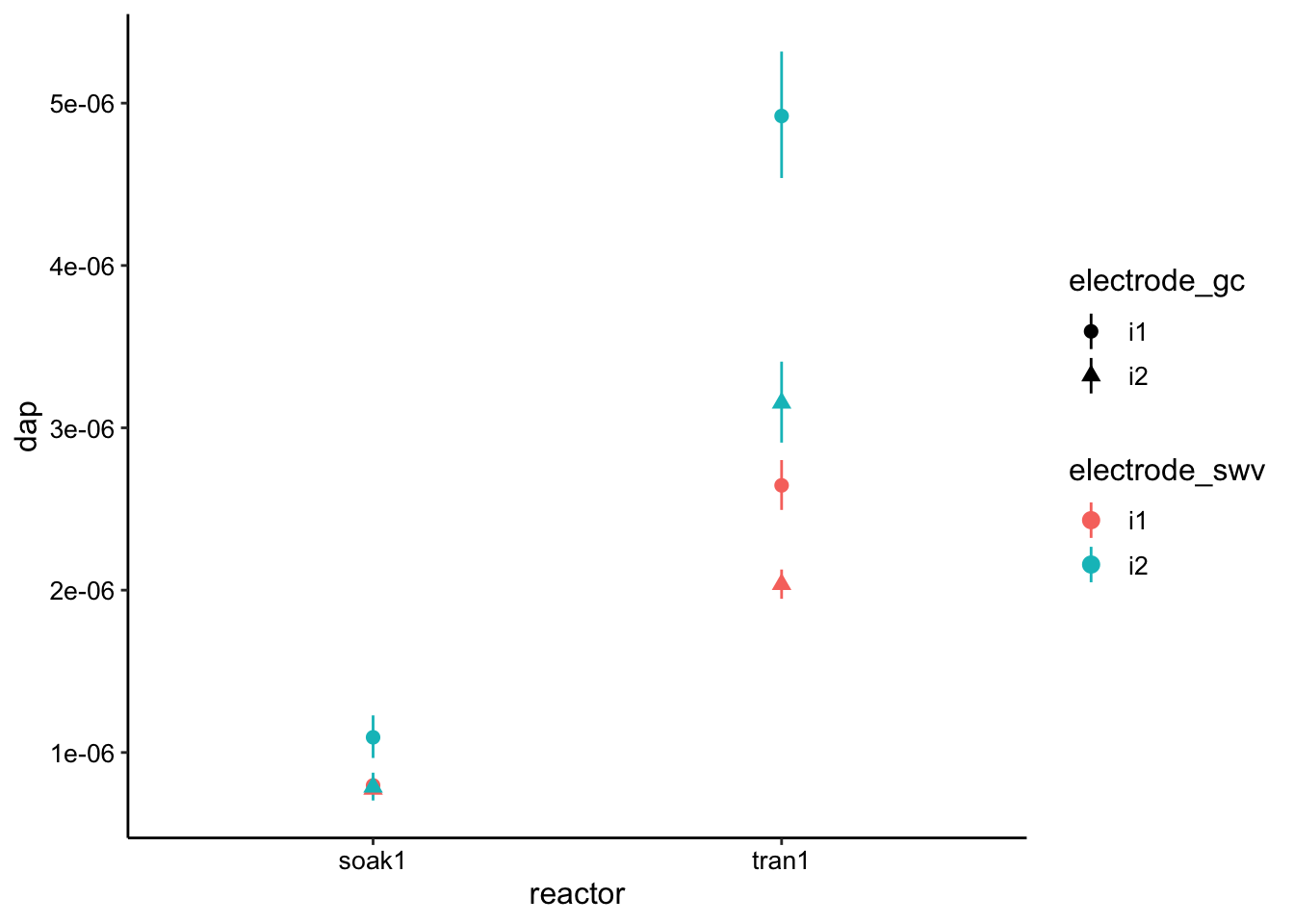
lms %>% kable() %>% kable_styling()
|
reactor
|
electrode_swv
|
electrode_gc
|
term
|
estimate
|
std.error
|
statistic
|
p.value
|
conf.low
|
conf.high
|
dap
|
dap_high
|
dap_low
|
dataset
|
|
soak1
|
i1
|
i1
|
max_current_swv
|
0.0679339
|
0.0003857
|
176.11233
|
0
|
0.0671629
|
0.0687050
|
8.0e-07
|
8.0e-07
|
8.0e-07
|
SWVvsGC
|
|
soak1
|
i1
|
i2
|
max_current_swv
|
0.0669625
|
0.0005771
|
116.02347
|
0
|
0.0658088
|
0.0681162
|
8.0e-07
|
8.0e-07
|
7.0e-07
|
SWVvsGC
|
|
soak1
|
i2
|
i1
|
max_current_swv
|
0.0795563
|
0.0023490
|
33.86869
|
0
|
0.0747826
|
0.0843300
|
1.1e-06
|
1.2e-06
|
1.0e-06
|
SWVvsGC
|
|
soak1
|
i2
|
i2
|
max_current_swv
|
0.0675213
|
0.0018069
|
37.36832
|
0
|
0.0638493
|
0.0711934
|
8.0e-07
|
9.0e-07
|
7.0e-07
|
SWVvsGC
|
|
tran1
|
i1
|
i1
|
max_current_swv
|
0.1237482
|
0.0017951
|
68.93727
|
0
|
0.1201563
|
0.1273402
|
2.6e-06
|
2.8e-06
|
2.5e-06
|
SWVvsGC
|
|
tran1
|
i1
|
i2
|
max_current_swv
|
0.1085590
|
0.0012016
|
90.34606
|
0
|
0.1061546
|
0.1109634
|
2.0e-06
|
2.1e-06
|
1.9e-06
|
SWVvsGC
|
|
tran1
|
i2
|
i1
|
max_current_swv
|
0.1687697
|
0.0032219
|
52.38178
|
0
|
0.1620878
|
0.1754515
|
4.9e-06
|
5.3e-06
|
4.5e-06
|
SWVvsGC
|
|
tran1
|
i2
|
i2
|
max_current_swv
|
0.1351031
|
0.0025761
|
52.44495
|
0
|
0.1297606
|
0.1404456
|
3.2e-06
|
3.4e-06
|
2.9e-06
|
SWVvsGC
|
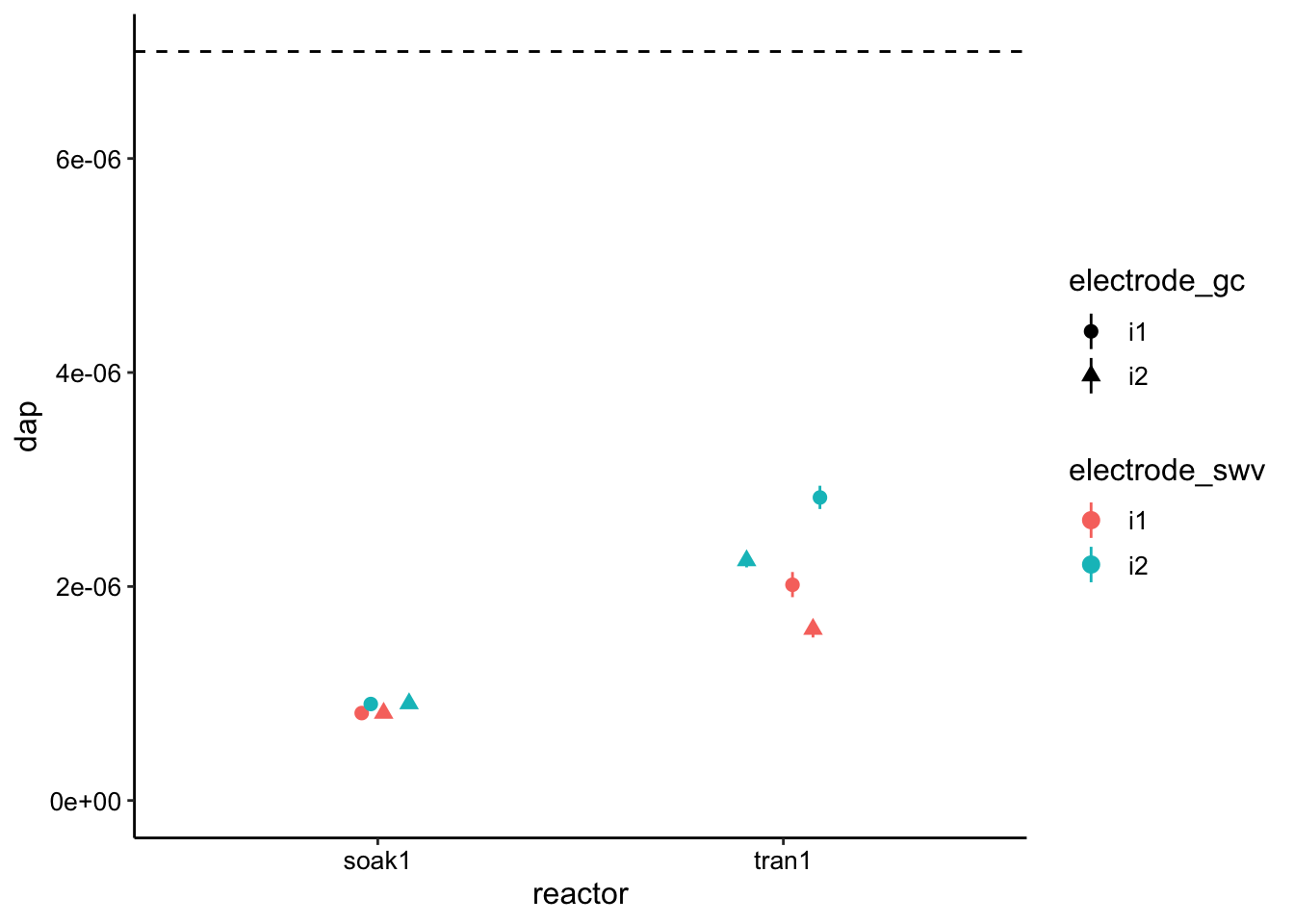
ggplot(lms, aes(x = reactor, y = dap, color = electrode_swv,
shape = electrode_gc)) + geom_pointrange(aes(ymin = dap_low,
ymax = dap_high))
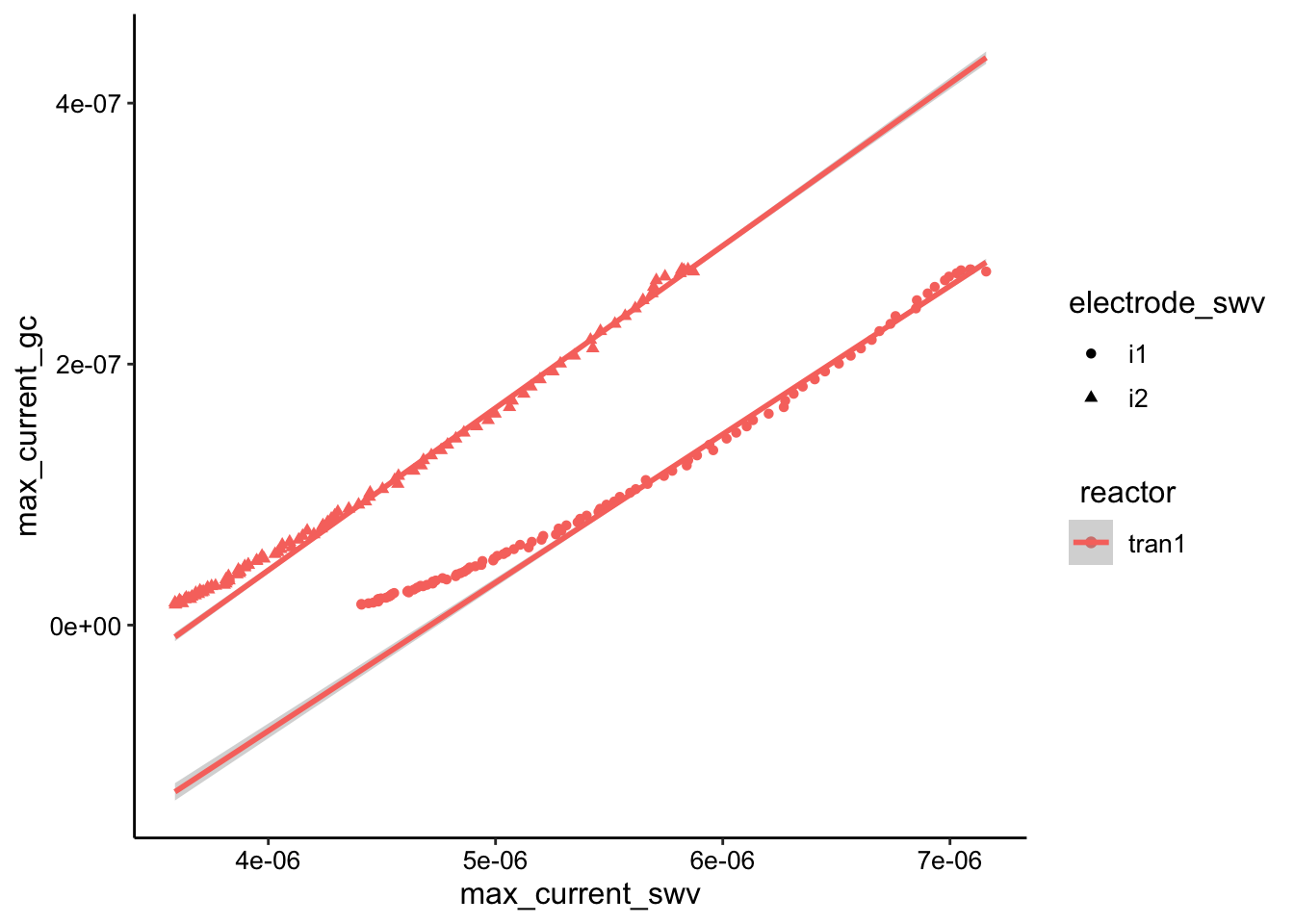
lm_soak <- swv_gc_max %>% filter(reactor == "soak1") %>% filter(rep >
0 & rep > 30) %>% group_by(reactor, electrode_swv, electrode_gc) %>%
do(tidy(lm(max_current_gc ~ max_current_swv, data = .), conf.int = T)) %>%
filter(term == "max_current_swv") %>% mutate(dap = dap_from_swvGC(m = estimate)) %>%
mutate(dap_high = dap_from_swvGC(m = conf.high)) %>% mutate(dap_low = dap_from_swvGC(m = conf.low)) %>%
mutate(dataset = "SWVvsGC")
lm_tran <- swv_gc_max %>% filter(reactor == "tran1") %>% filter(rep >
0 & max_current_swv < 50) %>% group_by(reactor, electrode_swv,
electrode_gc) %>% do(tidy(lm(max_current_gc ~ max_current_swv,
data = .), conf.int = T)) %>% filter(term == "max_current_swv") %>%
mutate(dap = dap_from_swvGC(m = estimate)) %>% mutate(dap_high = dap_from_swvGC(m = conf.high)) %>%
mutate(dap_low = dap_from_swvGC(m = conf.low)) %>% mutate(dataset = "SWVvsGC")
bind_rows(lm_soak, lm_tran) %>% ggplot(., aes(x = reactor, y = dap,
color = electrode_swv, shape = electrode_gc)) + geom_hline(yintercept = 7e-06,
linetype = 2) + geom_pointrange(aes(ymin = dap_low, ymax = dap_high),
position = position_jitter(width = 0.1, height = 0)) + ylim(0,
NA)